
KEY ACHIEVEMENTS | BACKGROUND | PRACTICE DEVELOPMENT | MEASURING IMPACT | KEY LEARNINGS
Key achievements
|
Background
The International Health Regulations (IHR) Strengthening Project provided lab-based training on whole genome sequencing to scientists at the Ethiopian Public Health Institute (EPHI) in Addis Ababa, from May 23rd - 26th 2022. This was an excellent example of how a training workshop originally developed for UKHSA Porton-based genome sequencing staff, could be adapted to work in labs in low-middle income countries (LMICs), one of the many benefits of intra-and inter-organisational collaboration. The lab-based course was delivered by members of the Novel and Dangerous Pathogens (NADP) Training team, on behalf of the UKHSA IHR project, and was supported by the UKHSA New Variant Assessment platform (NVAP) programme.

Practice Development
Six scientists, who will operate the newly established Pathogen Genomic Sequencing Core Facility (PGCF) at EPHI, were selected from different departments including the Polio reference lab, Influenza and arbovirus reference lab, HIV reference lab, and entomology, parasitology and bacteriology labs to implement sequencing capacity in those areas.
The in-country training was led by Prof. Christopher Logue, IHR Laboratory Capacity Building Lead for Ethiopia and International Training Lead for NADP Training, with support from fellow NADP Training trainer Dr Romisa Asadi. Bioinformatics training content was provided by Dr Karen Osman of the Genomics of Rare and Emerging Human Pathogens group, with remote bioinformatics support provided by Drs Babak Afrough, Sam Sims and Kate Edington of the NVAP bioinformatics team. The UKHSA Ethiopia team, Constantina Laou (NVAP) and Hiwot Hailu (IHR) participated as observers, and an invitation to observe was also accepted by the US CDC Ethiopia office.

Good pipetting to improve accuracy and reducing possible cross-contamination between samples.
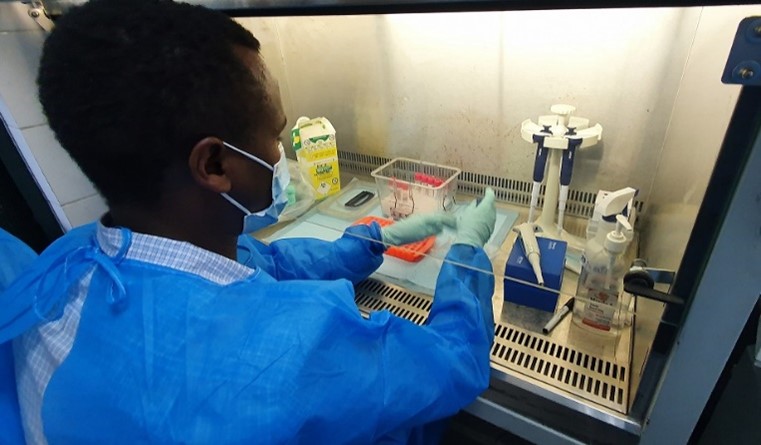
Inactivation of SARS-CoV-2 samples safely formed an important part of the laboratory training
The focus of the lab-based training was to build capability, competence and confidence in the end-to-end processing confirmed SARS-CoV-2 samples, from arrival in the lab to sequencing on an Oxford Nanopore Technologies (ONT) minION sequencing device. An additional aim of the work was to train participants on exporting the sequence to the SARS-CoV-2 Artic pipeline for analysis and subsequent sharing globally via GISAID.
This training complemented an earlier week-long remote training provided to EPHI PGCF staff by NVAP in collaboration with the University of Cambridge.

Discussing the most appropriate workflow to fit the existing laboratory footprint with the Sequencing laboratory manager and US CDC laboratory support team
Measuring Impact
The face-to-face laboratory training was very well received by all trainees and facilitators, it was summed up well by one of the participants:
“I enjoyed the course, it is easy, understandable and focuses on practice. This is what we want. The approach to deliver the course is fascinating. Within a short period of time we were introduced to all of the information/requirements for WGS using ONT, from safety to data analysis.”
Three of the seven suspect SARS-CoV-2 samples used for the training, had a sufficiently high enough quantities of virus present to allow for enough nucleic acid to be amplified and purified for sequencing.
As a result of the training these three samples produced the first ever SARS-CoV-2 sequences generated to date using an ONT minION in Ethiopia. Over 150 have been subsequently processed following the workshop on May 26th.

Knowledge check following sequence library preparation
Key Learnings
Follow up remote training in monkeypox detection by PCR has now also been developed by the NADP training team in collaboration with the Rare and Imported Pathogens Lab (RIPL) team and was delivered in EPHI, by Hiwot Hailu. This included the emergency provision of diagnostic reagents for testing suspect clinical samples. Follow up monkeypox WGS is planned and hands-on in-Country training in processing of high consequence pathogens for molecular detection is scheduled for September 2022, following a Laboratory Leaders’ workshop at EPHI in July.
By Dr Christopher Logue